

Research on Bioinformatics
Decoding Gene Expression using DNA Sequence and Exploiting Post-Translational Modifications
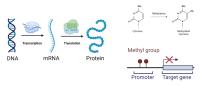
Understanding gene expression is crucial for deciphering cellular processes and disease mechanisms. Traditional methods primarily focus on the analysis of DNA sequence data to predict gene expression levels. However, recent research has highlighted the importance of post-translational modifications (PTMs) in regulating gene expression. PTMs, such as methylation, play key roles in modulating protein activity and stability, ultimately impacting gene expression.
By incorporating PTM data, researchers can gain a deeper understanding of the regulatory mechanisms governing gene expression dynamics. Our aim is to encompass computational and experimental approaches that leverage DNA sequence data and PTM profiles to predict and interpret gene expression patterns. Decoding gene expression through the synergy of DNA sequence analysis and PTM exploitation provides a powerful framework for unraveling the complexities of gene regulation and its impact on cellular function and disease processes.
Integrating Genomics and Transcriptomics: Advancements through Multi-View Approaches
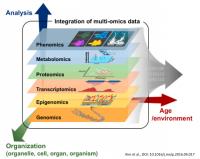
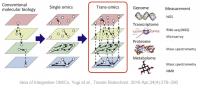
The integration of genomics and transcriptomics has revolutionized the understanding of biological systems by providing comprehensive insights into the relationship between genetic information and gene expression patterns. Traditionally, genomics and transcriptomics have been studied independently. Still, recent advancements in multi-view approaches have enabled the simultaneous analysis of genomic and transcriptomic data, leading to a more holistic understanding of cellular processes. Our main interests concern multi-view methods leveraging the complementary information encoded in genomic and transcriptomic datasets to uncover novel biological insights. Computational and statistical techniques employed in multi-view integration involve co-expression network analysis, clustering algorithms, and dimensionality reduction methods. The benefits of multi-view integration include the identification of regulatory mechanisms, the discovery of novel biomarkers, and the elucidation of complex gene regulatory networks. Integrating genomics and transcriptomics through multi-view approaches represents a powerful paradigm shift in biological research, enabling comprehensive analyses and opening new avenues for discoveries at the intersection of genetic variation and gene expression.
RNA and DNA-Based Gene Fusion Detection: Unveiling Genomic Rearrangements
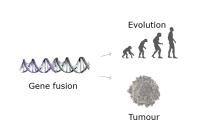
Gene fusions, resulting from genomic rearrangements, have emerged as important drivers of oncogenesis and potential therapeutic targets in various types of cancer. Detecting gene fusions accurately is crucial for understanding tumor biology and developing targeted therapies. In recent years, advancements in RNA and DNA-based approaches have significantly improved gene fusion detection capabilities.
Our aim is to unravel genomic rearrangements through RNA and DNA-based gene fusion detection tools, which are promising for advancing precision medicine in oncology and improving patient outcomes.
Predicting miRNA Targets in Humans: Computational Prediction Approaches
MicroRNAs (miRNAs) are small non-coding RNA molecules that play critical roles in post-transcriptional gene regulation. Identifying the targets of miRNAs is crucial for understanding their functional implications in various biological processes and diseases. Computational prediction approaches have emerged as valuable tools for predicting human miRNA targets, as experimental methods can be laborious and time-consuming. We are interested in computational prediction approaches for human miRNA target identification, providing algorithms and databases that leverage sequence complementarity and evolutionary conservation principles to predict miRNA-target interactions. As computational prediction approaches continue to improve, they provide valuable insights into miRNA target interactions, enhancing our understanding of gene regulation and the intricate mechanisms underlying human biology and disease.
Advancing Medulloblastoma Research: Genomics and Proteomics Insights from Sample Analysis
Medulloblastoma is a malignant brain tumor primarily affecting children, necessitating continuous advancements in research to improve diagnosis, prognosis, and treatment strategies. In recent years, integrating genomics and proteomics has emerged as a powerful approach for gaining comprehensive insights into the molecular landscape of medulloblastoma. Medulloblastoma research has made significant strides toward personalized medicine approaches and targeted therapies by leveraging genomics and proteomics insights from sample analysis. However, challenges remain, including data integration, validation, and translation into clinical practice.
This project is developed with the “Signaling in the Development and Brain Tumor” Lab at the Institut Curie Center, Orsay, Paris. Nonetheless, the advancements achieved through genomics and proteomics analyses of medulloblastoma samples have propelled the field forward, bringing us closer to improved patient outcomes and, ultimately, a cure for this devastating disease.
Unleashing the Power of OncoDash: Software for Informed Clinical Decisions

The increasing amounts of biological and patient clinical data and the rapid pace of associated research results offer new opportunities for improved treatment decisions but also increase the complexity of the decision-making process. The Oncodash software addresses this problem by assisting clinicians with patient review and the prioritization of treatment options. OncoDash provides interactive interfaces on not only patient data but also a rich environment to interact with models. In particular, it includes AI-driven models, which show surprising levels of performance in health and across other applications.
A Convergence of Histopathology and Genetics for Anticipating Chemotherapy Response in ovarian cancer.
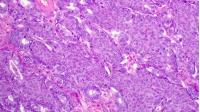
Recent advancements in histopathology and genetics have enabled the development of personalized medicine approaches that can improve patient outcomes by tailoring treatments based on individualized genetic and clinical characteristics. In the field of oncology, chemotherapy is often used as a treatment option, but the efficacy of chemotherapy varies between patients due to differences in genetic makeup and tumor characteristics. Therefore, identifying predictive biomarkers of chemotherapy response is crucial for effective treatment planning. In this context, we integrate histopathology images with genetic features aimed at predicting chemotherapy response in ovarian cancer patients. Ultimately, the convergence of histopathology and genetics has the potential to transform cancer treatment by enabling the development of more personalized and effective treatment plans based on an individual patient's unique molecular profile.
Studying host-pathogen interaction via microscopy and Deep Learning: application to antimicrobial resistant bacteria and monoclonal antibodies discovery

This research activity aims to develop deep-learning techniques for analyzing biological images of the Opsonophagocytosis assay. This critical immune process involves antibodies marking pathogens for destruction by phagocytes.
Identification of AI-Based Biomarkers to Predict HCC Response to Medical Treatment

Hepatocellular Carcinoma (HCC) is one of the most common and deadly forms of liver cancer: early diagnosis and effective treatment planning are critical to improving survival rates.
A key challenge in managing HCC is accurately predicting how individual patients will respond to different medical treatments, such as Immune Checkpoint Inhibitors (ICI) and Anti-Angiogenic (AA) therapies. The effectiveness of these treatments varies greatly among patients, underscoring the importance of personalized treatment strategies for improving patient outcomes.
In collaboration with Humanitas Research Hospital, we focus on leveraging advanced AI techniques, particularly Deep Learning, to identify imaging biomarkers that can help differentiate between patients based on their prognosis and likelihood of responding to specific therapies.
Multi-Omic Graph Neural Networks for Cancer Prognosis and Biomarker Identification
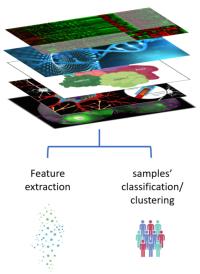
Cancer is a multifaceted and heterogeneous disease, posing challenges in predicting its outcome due to the intricate interplay of various molecular factors. Understanding the influence of diverse biological data, including mRNA expression, DNA methylation, and copy number variations (CNVs), is crucial for enhancing prognosis and treatment strategies in cancer progression. These varied molecular data types yield valuable insights into gene regulation, genetic instability, and tumor growth, offering complementary perspectives on cancer biology.
Multimodal Deep Learning Model for cancer prediction

Pathologists who specialize in studying histopathological tumor images play a crucial role in personalized cancer treatment. By examining a tumor tissue samples, they analyze the structure, composition, and behavior of cancer cells. This process helps identify the specific type of tumor, its grade, and its potential aggressiveness. Their work goes beyond basic diagnosis, as they also identify molecular and genetic markers that provide insights into how a particular tumor may respond to different treatments. These insights allow oncologists to tailor therapies to the individual characteristics of a patient’s cancer, leading to more effective and targeted treatment plans. This personalized approach helps improve patient outcomes, reduces unnecessary side effects, and ensures that treatments are more aligned with the biological profile of the tumor. The precision and expertise of pathologists are invaluable in the evolving field of precision medicine, where cancer treatments are increasingly based on a deep understanding of the tumor's unique biology.