Predicting miRNA Targets in Humans: Computational Prediction Approaches
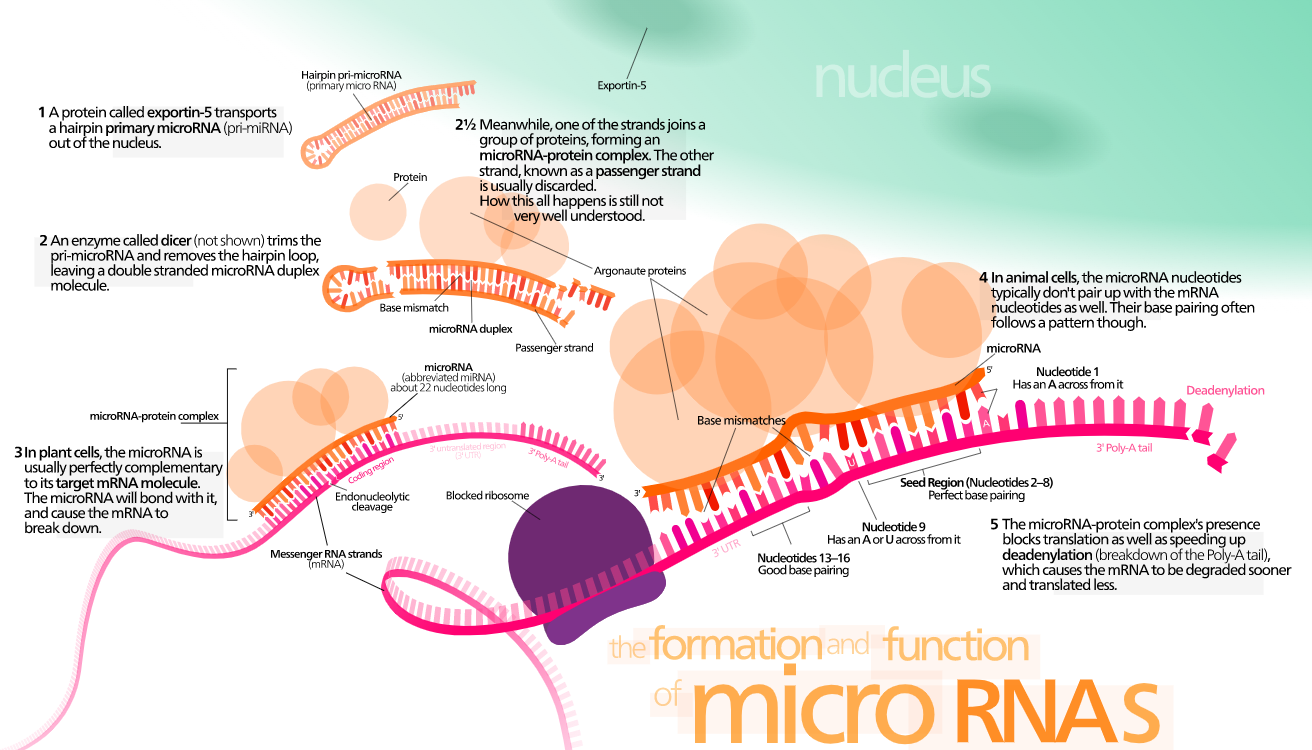
MicroRNAs (miRNAs) are small non-coding RNA molecules that play critical roles in post-transcriptional gene regulation. Identifying the targets of miRNAs is crucial for understanding their functional implications in various biological processes and diseases. Computational prediction approaches have emerged as valuable tools for predicting human miRNA targets, as experimental methods can be laborious and time-consuming. We are interested in computational prediction approaches for human miRNA target identification, providing algorithms and databases that leverage sequence complementarity and evolutionary conservation principles to predict miRNA-target interactions. As computational prediction approaches continue to improve, they provide valuable insights into miRNA target interactions, enhancing our understanding of gene regulation and the intricate mechanisms underlying human biology and disease.
This research area aims to reveal pathology-determining microRNA targets in multi-omic datasets. In detail, datasets comprise miRNA, mRNA, and proteomics data related to the same patients. The main computational contribution to the field consists of designing and implementing algorithms based on the statistical conditional probability and ML/DL techniques to infer the impact of miRNA post-transcriptional regulation on target genes exploiting the protein expression values. These methodologies allow a more in-depth understanding and identification of target genes.