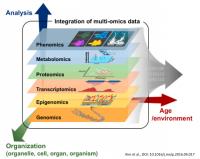
Integrating Genomics and Transcriptomics: Advancements through Multi-View Approaches
The integration of genomics and transcriptomics has revolutionized the understanding of biological systems by providing comprehensive insights into the relationship between genetic information and gene expression patterns. Traditionally, genomics and transcriptomics have been studied independently. Still, recent advancements in multi-view approaches have enabled the simultaneous analysis of genomic and transcriptomic data, leading to a more holistic understanding of cellular processes. Our main interests concern multi-view methods leveraging the complementary information encoded in genomic and transcriptomic datasets to uncover novel biological insights. Computational and statistical techniques employed in multi-view integration involve co-expression network analysis, clustering algorithms, and dimensionality reduction methods. The benefits of multi-view integration include the identification of regulatory mechanisms, the discovery of novel biomarkers, and the elucidation of complex gene regulatory networks. Integrating genomics and transcriptomics through multi-view approaches represents a powerful paradigm shift in biological research, enabling comprehensive analyses and opening new avenues for discoveries at the intersection of genetic variation and gene expression.
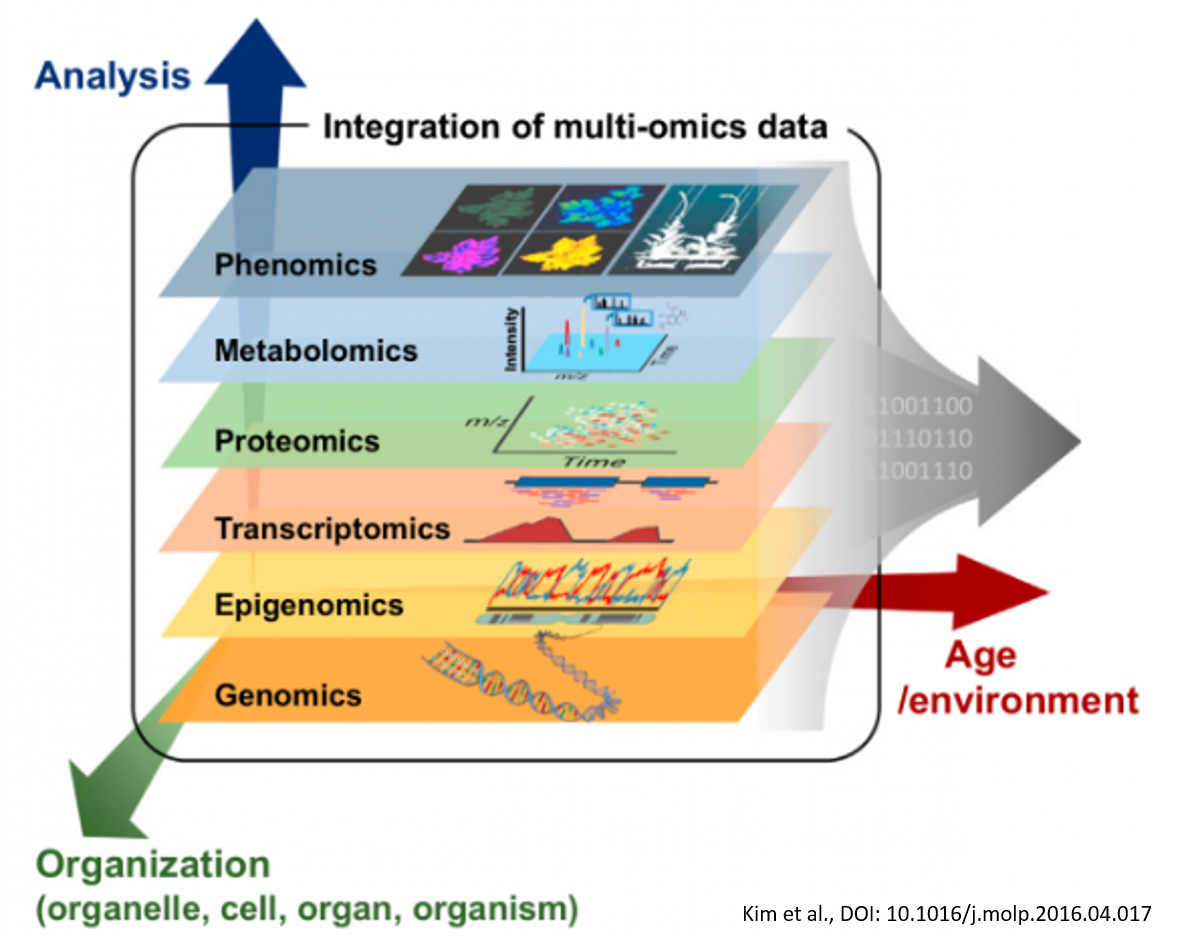
Due to the continuous increase in the number and complexity of genomics and biological data, new computer science techniques are needed to analyze these data and provide valuable insights into the main features.
Consequently, many heterogeneous complex data related to several diseases and healthy individuals are now available (e.g., RNA-seq, gene expression data, miRNAs expression data, methylation sequencing data, and many others). Each of these data is also called omic, and their integrative study is called multi-omics. In this context, the aim is to integrate multi-omics data involving thousands of features (genes, microRNA) and identify which are relevant for a specific biological process.
From a computational point of view, finding the best strategies for multi-omics analysis and relevant feature identification is a very open challenge.
Publications
| 1 |
Lovino, Marta; Randazzo, Vincenzo; Ciravegna, Gabriele; Barbiero, Pietro; Ficarra, Elisa; Cirrincione, Giansalvo
"A survey on data integration for multi-omics sample clustering"
NEUROCOMPUTING,
vol. 488,
pp. 494
-508
,
2022
| DOI: 10.1016/j.neucom.2021.11.094
Journal

|
| 2 | Barbiero, Pietro; Lovino, Marta; Siviero, Mattia; Ciravegna, Gabriele; Randazzo, Vincenzo; Ficarra, Elisa; Cirrincione, Giansalvo "Unsupervised Multi-Omic Data Fusion: the Neural Graph Learning Network" LNCS, 16th International Conference on Intelligent Computing, ICIC 2020, vol. 12463, Bari (ita), pp. 172 -182 , Ottobre 2020, 2020 | DOI: 10.1007/978-3-030-60799-9_15 Conference |
| 3 | Lovino, Marta; Bontempo, Gianpaolo; Cirrincione, Giansalvo; Ficarra, Elisa "Multi-omics classification on kidney samples exploiting uncertainty-aware models" LNCS, vol. LNCS, Bari (Online), pp. 32 -42 , Oct 10-11th 2020, 2020 | DOI: 10.1007/978-3-030-60802-6_4 Conference |
| 4 |
Roberti, Ilaria; Lovino, Marta; Di Cataldo, Santa; Ficarra, Elisa; Urgese, Gianvito
"Exploiting Gene Expression Profiles for the Automated Prediction of Connectivity between Brain Regions"
INTERNATIONAL JOURNAL OF MOLECULAR SCIENCES,
vol. 20,
pp. N/A
-N/A
,
2019
| DOI: 10.3390/ijms20082035
Journal

|