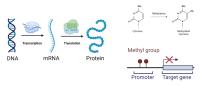
Decoding Gene Expression using DNA Sequence and Exploiting Post-Translational Modifications
Understanding gene expression is crucial for deciphering cellular processes and disease mechanisms. Traditional methods primarily focus on the analysis of DNA sequence data to predict gene expression levels. However, recent research has highlighted the importance of post-translational modifications (PTMs) in regulating gene expression. PTMs, such as methylation, play key roles in modulating protein activity and stability, ultimately impacting gene expression.
By incorporating PTM data, researchers can gain a deeper understanding of the regulatory mechanisms governing gene expression dynamics. Our aim is to encompass computational and experimental approaches that leverage DNA sequence data and PTM profiles to predict and interpret gene expression patterns. Decoding gene expression through the synergy of DNA sequence analysis and PTM exploitation provides a powerful framework for unraveling the complexities of gene regulation and its impact on cellular function and disease processes.
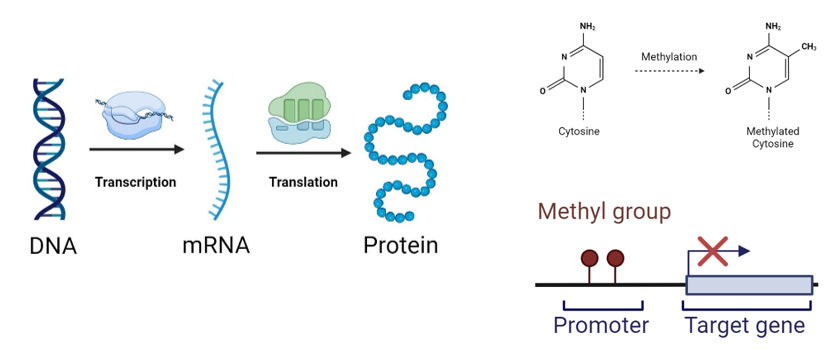
Gene expression is fundamental for understanding cellular processes and the mechanisms underlying diseases. It reflects how the information encoded in DNA is translated into functional proteins, directly influencing cellular and organismal health.
Traditional methods for predicting gene expression primarily focus on analyzing DNA sequence data, but they do not fully capture the complex regulatory dynamics of gene activity.
DNA methylation is an epigenetic mechanism that influences gene expression, playing a crucial role in regulating gene activity. This modification affects transcription without altering the DNA sequence and is strongly associated with diseases like cancer. The promoter region regulates gene activation; when a methyl group is added, it can silence a gene by preventing its transcription. This study aims to enhance gene expression prediction by integrating DNA sequence and methylation data. By leveraging computational approaches and deep learning models, we analyze the synergy between these two data sources, revealing the regulatory mechanisms governing gene expression.
Our investigation indicates that incorporating methylation features significantly enhances the predictive power of sequence models. This combined analysis provides a robust framework for deciphering the complexities of gene regulation and its implications for cellular functions and pathological processes. Furthermore, our findings deepen the understanding of the molecular mechanisms underlying gene regulation and hold promise for clinical applications, paving the way for personalized therapies and targeted interventions. Utilizing artificial intelligence models allows us to unveil intricate biological patterns and interpret how variations in methylation influence gene expression, resulting in improved insights into pathologies and enhanced clinical management.
Publications
| 1 |
Pipoli, Vittorio; Cappelli, Mattia; Palladini, Alessandro; Peluso, Carlo; Lovino, Marta; Ficarra, Elisa
"Predicting gene expression levels from DNA sequences and post-transcriptional information with transformers"
COMPUTER METHODS AND PROGRAMS IN BIOMEDICINE,
vol. 225,
pp. 107035
-107044
,
2022
| DOI: 10.1016/j.cmpb.2022.107035
Journal

|